Explicator is a C++ string translation library that uses a combination of string similarity metrics to provide fuzzy matching. The included metrics are
See here for more information.
Computer programs have trouble interpreting data intended for humans. Humans try to get around this problem by coming up with conventions, languages, and binary formats to capture meaning for a subset of data. This does not solve the problem for good, just lets us mostly get on with our work. Such behaviour is illustrated by the creation of large databases filled with information which is distilled from information-rich, readily-available sources.
This idea is analogous to Yahoo! in the early days of the internet. Yahoo attempted to classify the web in the form of collections of logical links. This approach was doomed to fail, mostly because the rise of online search (e.g. Google) greatly out-performed link aggregators. Both in terms of performance and relevancy. Google could automatically distill data down to something the users asked for.
I’ve written two software libraries (Explicator and Demarcator) that address this issue in a specific domain: radiotherapy patient data. Instead of attempting to classify and aggregate patient data into logical links, these libraries let users interpret data as it is encountered – no SQL schemas or prepared layouts, just ad-hoc, on-the-fly interpretation.
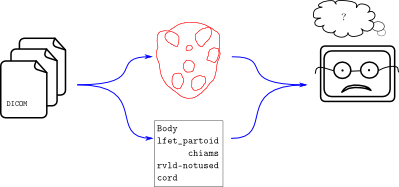
Contour data is noisy. Humans have all sorts of naming schemes for things. We don’t like to type out “Left Parotid” and so instead invent clever nicknames like “l_par” or “lfet partoid” (sic). When a researcher needs to process thousands of patient files, it becomes impractical to deal manually with the clever names in each data set. This is the problem that Explicator and Demarcator can solve. Here’s a workflow diagram showing where these libraries fit in:

The flow of data from a DICOM file – geometrical and lexicographical contour data, in this case – is unstructured and the analysis program on the right isn’t able to cope. It’s a one-off program which performs some wonderfully complex dosimetric computations. It just can’t handle things like “lfet partoid”. And it shouldn’t! This would be added complexity in an already complicated program. Instead, it should use a compatibility layer like Explicator or Demarcator.
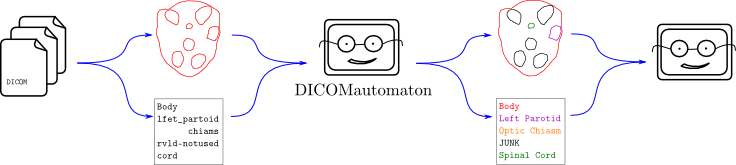
I believe the ideal solution is to ask the user to provide domain knowledge up front, draw as much information from that domain knowledge as reasonably possible, provide an easy library interface to use that knowledge for translation of messy data, and try improve translation as additional information becomes available. This is what Explicator and Demarcator were designed to do.
If Explicator or Demarcator are used, the one-off program above will effectively be able to read any DICOM-format data given to it without having to explicitly be programmed to perform any nitty-gritty translations. In a nutshell, Explicator and Demarcator are like a fuzzy Rosetta Stone for radiotherapy applications. If somebody, somewhere along the line accidentally puts a “lfet partoid” into the data, the one-off program will easily be able to handle it. In fact, the translation filter means the “lfet partoid” will be completely hidden from the one-off program. The only thing it will encounter is a nicely-formatted “Left Parotid”.
Both Explicator and Demarcator are part of the DICOMautomaton software suite. They are designed specifically to identify unknown contoured structures in DICOM files. Explicator works on contour labels whereas Demarcator works on 3D structures (volumes). They are both written in C++, and are both GPLv3 software.
Explicator (the contour name recognizer) is available on here. The Explicator approach is lightweight and generally works quite well. Demarcator (the geometrical tool) works better in some cases, but requires drastically more storage space and computation time. The code may be released in the future. Please enquire here.
Please send questions, comments, and pull requests here.